In this project, we will build a powerful binary image classifier, to distinguish COVID-19 viral pneumonia chest X-rays from non COVID-19 viral pneumonia chest X-rays. Before we begin, let me emphasize that this is not a diagnostic tool. The intent of this project is to showcase how AI can be useful in assisting COVID-19 detection. In our set up we will:
- retrieve images for COVID-19 chest X-rays and non COVID-19 viral pneumonia chest X-rays
- create training, validation and testing datasets
- use a pretrained deep neural network to train and fine tune to build a chest X-ray classifier
Dataset Details
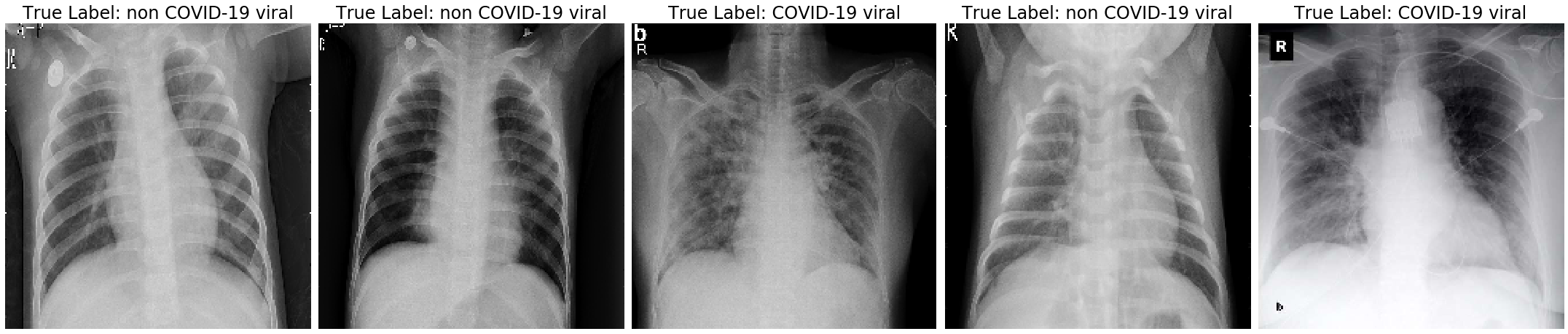
For this project we can collect and combine data from two different sources. Non COVID-19 viral pneumonia chest X-rays can be retrieved from the Kaggle website. X-rays in this dataset are from healthy individuals and from patients with both viral and bacterial infections. Using the information stored in the filenames we can retrieve 1314 viral pneumonia X-rays (non COVID-19). COVID-19 chest X-rays are released under the following URL: https://github.com/ieee8023/ covid-chestxray-dataset. Additional information about this dataset is available in the paper published by Cohen et.al. At the time of this project ~191 COVID-19 X-rays are available in this dataset. We will set aside ~ 30 images from each class as test data to evaluate our model leaving us with only 161 images to learn COVID-19 specific features and build a classifier.
Convolutional neural networks (Convnets/CNNs)
Convolutional neural networks (Convnets/CNNs) are the right tool for this job. Convnets are very good at learning features automatically from the training data and distinguish objects based on features learned. Applications of CNNs range widely, from speech recognition, face recognition, or traffic sign classification. In the context of medical imaging, CNNs have been used to classify medical images, detect cancer tissue in histopathological images, extract prognosticators from tissue microarrays (TMAs) of human solid tumors, and classify tumor cell nuclei according to chromatin patterns. Convnets perform reasonably well even on small datasets without custom feature engineering, but even with few hundred images it is a very difficult task to build and train models that generalize well.
Data Augmentation and Transfer Learning (Mitigating problems with small data size)
To mitigate problems commonly seen with deep neural networks trained with small datasets such as generalization and difficulty in reaching the global optimum we will employ data augmentation and transfer learning.
Data Augmentation
To make most out of these limited data samples, we will perform a number of random image transformations and increase the COVID-19 data size to 600 images. We will use imgaug which is a python library to augment training data. During the training phase we will load the dataset and perform real-time augmentation with Keras ImageDataGenerator class so the network sees different images at each epoch. We will not apply any transformations on our test data except scaling the images (which is mandatory).
Let's look at an example right away:
sometimes = lambda aug: iaa.Sometimes(0.5, aug)
apply the following augmenters to most images
seq = iaa.Sequential([
iaa.Fliplr(0.5), # horizontally flip 50% of all images
iaa.Flipud(0.2), # vertically flip 20% of all images
sometimes(iaa.Affine(
translate_percent={"x": (-0.2, 0.2), "y": (-0.2, 0.2)}, # translate by -20 to +20 percent (per axis)
rotate=(-25, 25), # rotate by -25 to +25 degrees
shear=(-8, 8), # shear by -8 to +8 degrees
order=[0, 1], # use nearest neighbour or bilinear interpolation (fast)
cval=(0, 255), # if mode is constant, use a cval between 0 and 255
mode=ia.ALL # use any of scikit-image's warping modes (see 2nd image from the top for examples)
)),
iaa.SomeOf((0, 5),
[iaa.OneOf([
iaa.GaussianBlur((0, 3.0)), # blur images with a sigma between 0 and 3.0
iaa.AverageBlur(k=(2, 7)), # blur image using local means with kernel sizes between 2 and 7
]),
iaa.Sharpen(alpha=(0, 1.0), lightness=(0.75, 1.5)), # sharpen images
iaa.AdditiveGaussianNoise(loc=0, scale=(0.0, 0.05*255), per_channel=0.5), # add gaussian noise to images
],
random_order=True
)],
random_order=True
)
def augment(img):
"""
to_deterministic() removes the randomness from all augmenters and makes them deterministic
(e.g. for each parameter that comes from a distribution, it samples one value from that
distribution and then keeps reusing that value)
"""
seq_det = seq.to_deterministic()
aug_img = seq_det.augment_image(img)
aug_img = vgg19_preprocess_input(aug_img)
return aug_img
# Instantiate the ImageDataGenerator from tensorflow.keras passing in out custom augmentation function
train_generator = ImageDataGenerator(preprocessing_function=augment)
for f in f_paths:
failed_files = []
try:
img = load_img(f)
x = img_to_array(img)
x = x.reshape((1, ) + x.shape)
i = 0
for batch in train_generator.flow(x, batch_size = 1,
save_to_dir ="./covid_aug",
save_prefix ='covid_aug', save_format ='jpeg'):
i += 1
if i > 5:
break
except:
failed_files.append(f)
Transfer Learning
The augmented images are highly correlated and the models trained on few hundred images can learn only limited number of features and have limited capacity to generalize well. So we can rely on models that were trained on large datasets. A large number of pretrained CNN models, all of which pretrained on diverse image categories on the ImageNet database are now publicly available for reuse. We will choose VGG19 in particular and repurpose the diverse set of features learned over a million images for the X-ray classification task at hand.
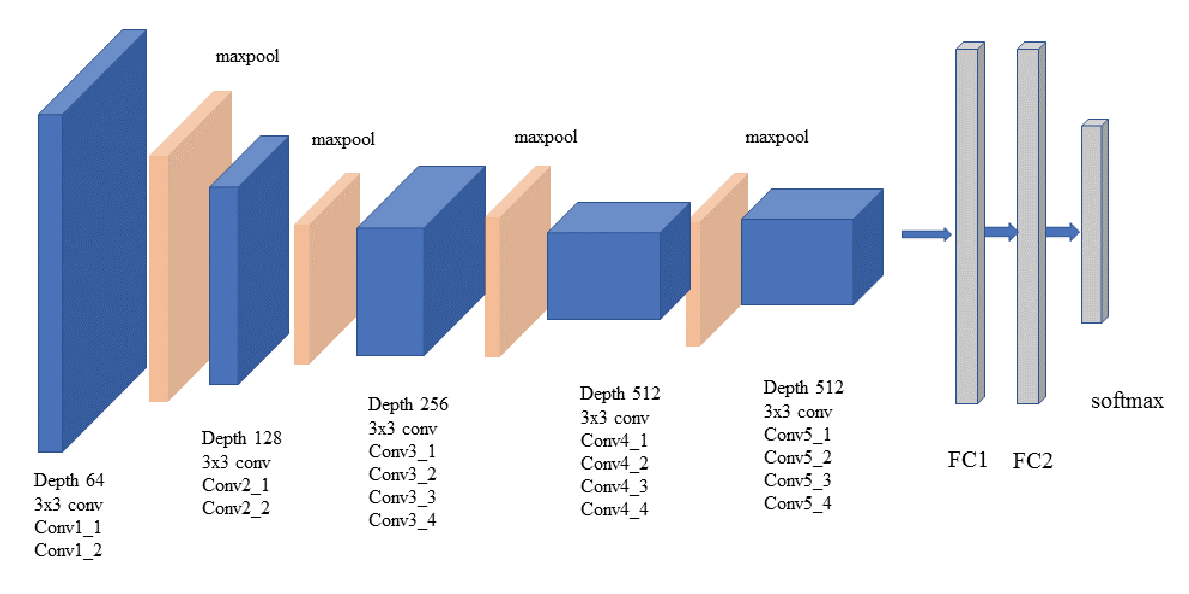
VGG19 Architecture
VGG19 is a 19 layer deep neural network with a simple architecture, a convolution base and a fully connected dense classifier as depicted below. The convolution base is built with (blue) 5 blocks of convolution layers separated by (tan) maxpooling layers (2X2 maxpooling with a stride of 2) for downsampling. All convolution blocks use 3X3 convolutions with “same” padding and a stride of 2. The output of the convolutions base is connected to three dense layers (FC1, FC2 and softmax). The dense layers are specific to the task the network was trained for.
Model 1: Pretrained model as Feature Extractor:
For our first model we will instantiate the pretrained VGG19 model up to the fully-connected layers with its pretrained weights. We will add a classifier initialized with random weights that will be trained for X-ray classification task. This classifier includes fully connected Dense, BatchNormalization and Dropout layers. During the training phase large gradient updates triggered by the randomly initialized weights would wreck the learned weights in the convolution base. To avoid any changes in the pretrained weights we will freeze the pretrained layers and train just the classifier.
# Instantiate the vgg19 model without the top classifier.
base_model = VGG19(input_shape=(224, 224, 3),
weights='imagenet', include_top=False)
# Add a classifier to the convolution block classifier
out = base_model.output
x = Flatten()(out)
# Add a classifier
x = Dense(1024, activation='relu', name='dense_01')(x)
x = BatchNormalization(name='batch_normalization_01')(x)
x = Dropout(0.5, name='dropout_01')(x)
x = Dense(512, activation='relu', name='dense_02')(x)
x = Dropout(0.3, name='dropout_02')(x)
output = Dense(2, activation='softmax', name='output')(x)
# Define the model
model_vgg19_binary_01 = Model(base_model.inputs, output)
# Freeze all layers in the convolution block. We don't want to train these weights yet.
for layer in base_model.layers:
layer.trainable = False
model_vgg19_binary_01.summary()
# Compile a model
model_vgg19_binary_01.compile(
loss="categorical_crossentropy", optimizer=RMSprop(0.001), metrics=["accuracy"])
filepath = "./output_3/aug_covid_binary_model_weights_ft.h5"
es = EarlyStopping(monitor='loss', verbose=1, mode='min', patience=4)
cp = ModelCheckpoint(filepath, monitor='val_loss', verbose=1, save_best_only=True,
save_weights_only=False, mode='auto', save_freq='epoch')
# Set the number of training and validation steps
STEP_SIZE_TRAIN = train_datagenerator.n//train_datagenerator.batch_size
STEP_SIZE_VALID = valid_datagenerator.n//valid_datagenerator.batch_size
# Model fitting
history_01 = model_vgg19_binary_01.fit_generator(train_datagenerator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_datagenerator,
validation_steps=STEP_SIZE_VALID,
callbacks=[es, cp],
class_weight=class_weights,
epochs=10)
We will train this model for 10 epochs and save the model. To evaluate the model performance let’s plot the model’s accuracy and loss curves between training and validation data and look at the Precision and Recall scores in the classification report .
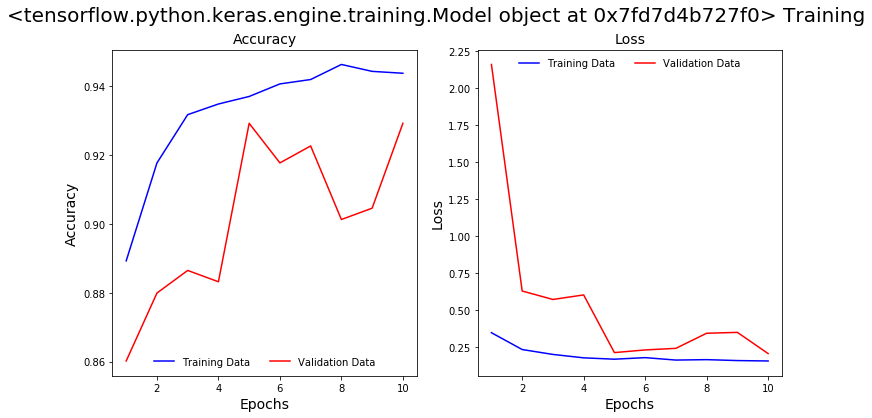
The plots show a validation accuracy of 94% which is pretty good. The accuracy and loss curves didn’t plateau and it appears that we can improve the model with more training.
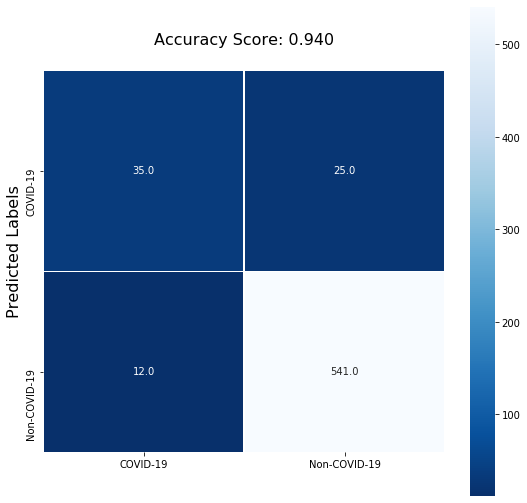
From the classification report on the validation data it appears that the model did not learn well: it captured only 58% of the total positive cases (recall) and only 74% of the COVID positive predictions are actually positive (precision).
Classification Report on Validation Data
| | precision |
recall |
f1-score |
support |
|---|---|---|---|---|
| COVID | 0.74 | 0.58 | 0.65 | 60 |
| non COVID | 0.96 | 0.98 | 0.97 | 553 |
| accuracy | 0.94 | 613 | ||
| macro avg | 0.85 | 0.78 | 0.81 | 613 |
| weighted avg | 0.94 | 0.94 | 0.94 | 613 |
Model 2: Fine tuning the convolution layers of a pre-trained network
Let’s train the model for more epochs but this time we will unfreeze the last convolution block (# 5) and fine tune it’s weights along with the classifier’s weights. We will construct the model as above and load the weights from previously trained model.
# # Define the model
model_vgg19_binary_02 = Model(base_model.inputs, output)
model_vgg19_binary_02.load_weights("./output_3/aug_covid_binary_model_weights_ft.h5")
# Freeze convolution block 1-4 i.e, upto layer 17.
for layer in base_model.layers[:17]:
layer.trainable = False
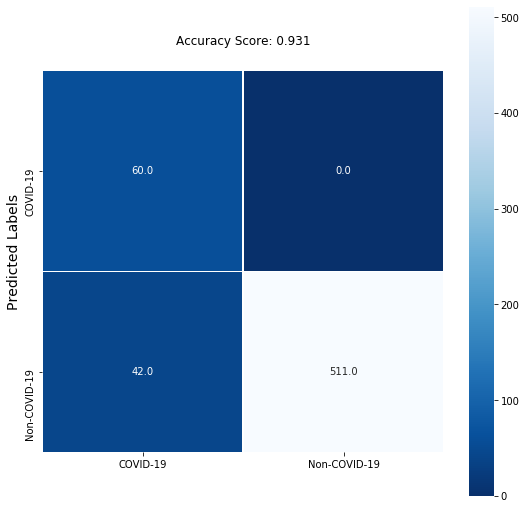
We will train this model for another 15 epochs using similar configurations and call backs and save the model. From the plots below we can see that the accuracy and loss continue to improve. The model successfully captured 100% of positive cases in the validation data but it incorrectly predicted a large number of positive predictions (false positive predictions).
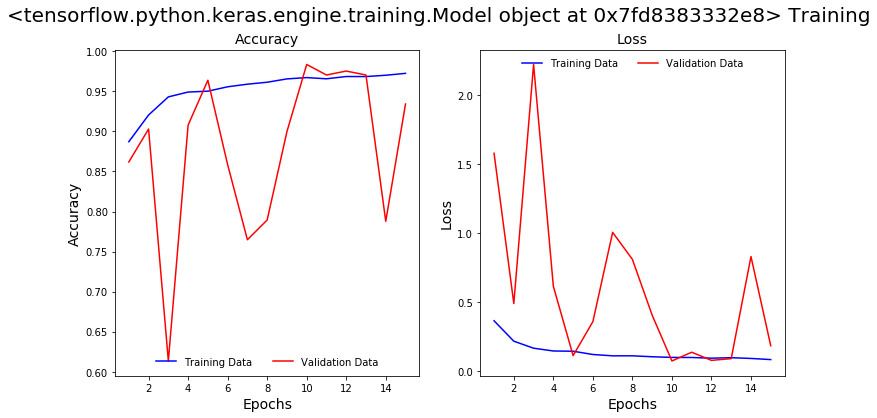
Classification Report on Validation Data
| | precision |
recall |
f1-score |
support |
|---|---|---|---|---|
| COVID | 0.59 | 1.00 | 0.74 | 60 |
| non COVID | 1.00 | 0.92 | 0.96 | 553 |
| accuracy | 0.93 | 613 | ||
| macro avg | 0.79 | 0.96 | 0.85 | 613 |
| weighted avg | 0.96 | 0.93 | 0.94 | 613 |
Model 3: Fine tuning the convolution layers of a pre-trained network
Now we will unfreeze another convolution block (# 4) and fine tune convolution blocks 4, 5 and the top classifier. We will instantiate a model as above and load the weights from model_02. This time we will also change the optimizer from RMSprop to Stochastic Gradient Descent (SGD) and train the model for another 25 epochs.
::: Python 3
# # Define the model
model_vgg19_binary_03 = Model(base_model.inputs, output)
model_vgg19_binary_03.load_weights("./output_3/aug_covid_binary_model_weights_02.h5")
# Freeze convolution block 1-4 i.e, upto layer 12.
for layer in base_model.layers[:12]:
layer.trainable = False
sgd = SGD(learning_rate=1e-4, decay=1e-6, momentum=0.9, nesterov=True)
# Compile a model
model_vgg19_binary_03.compile(
loss="categorical_crossentropy", optimizer=sgd, metrics=["accuracy"])
The model accuracy and loss stopped improving after 17 epochs of training and the model training was terminated by our EarlyStopping callback.
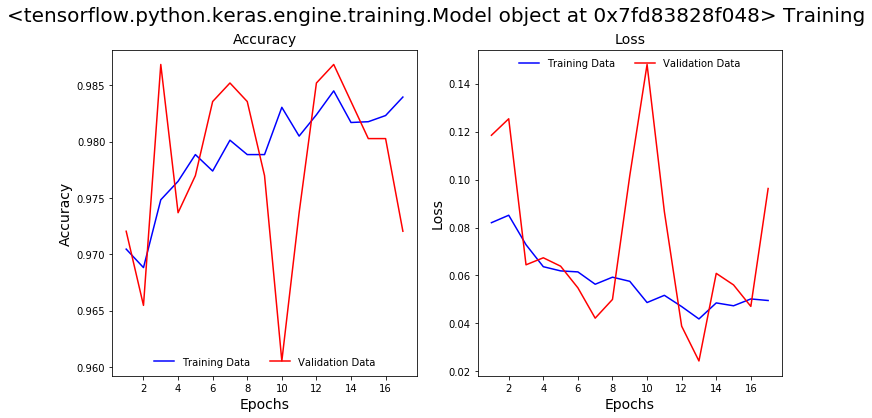
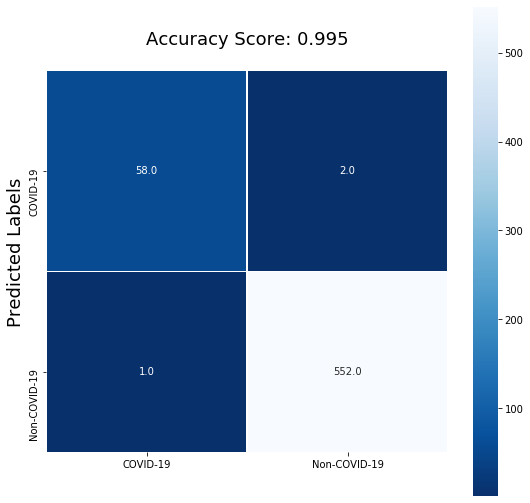
Both the Recall and Precision of the model on the validation data improved significantly. The model captured 97% of the COVID-19 positive samples in our validation data and 98% of the positive predictions are true positives.
Classification Report on Validation Data
| | precision |
recall |
f1-score |
support |
|---|---|---|---|---|
| COVID | 0.98 | 0.97 | 0.97 | 60 |
| non COVID | 1.00 | 1.00 | 1.00 | 553 |
| accuracy | 1.00 | 613 | ||
| macro avg | 0.99 | 0.98 | 0.99 | 613 |
| weighted avg | 1.00 | 1.00 | 1.00 | 613 |
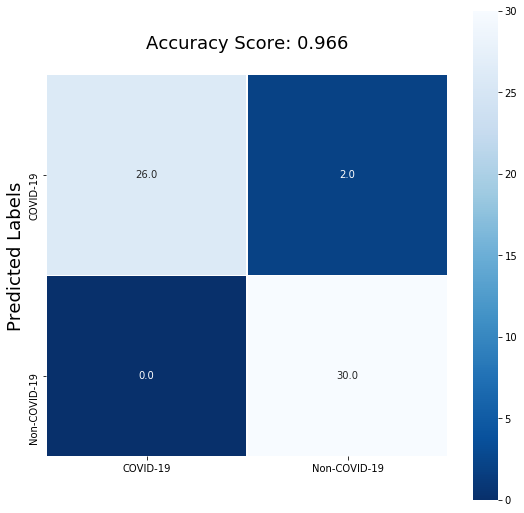
Evaluation on test images
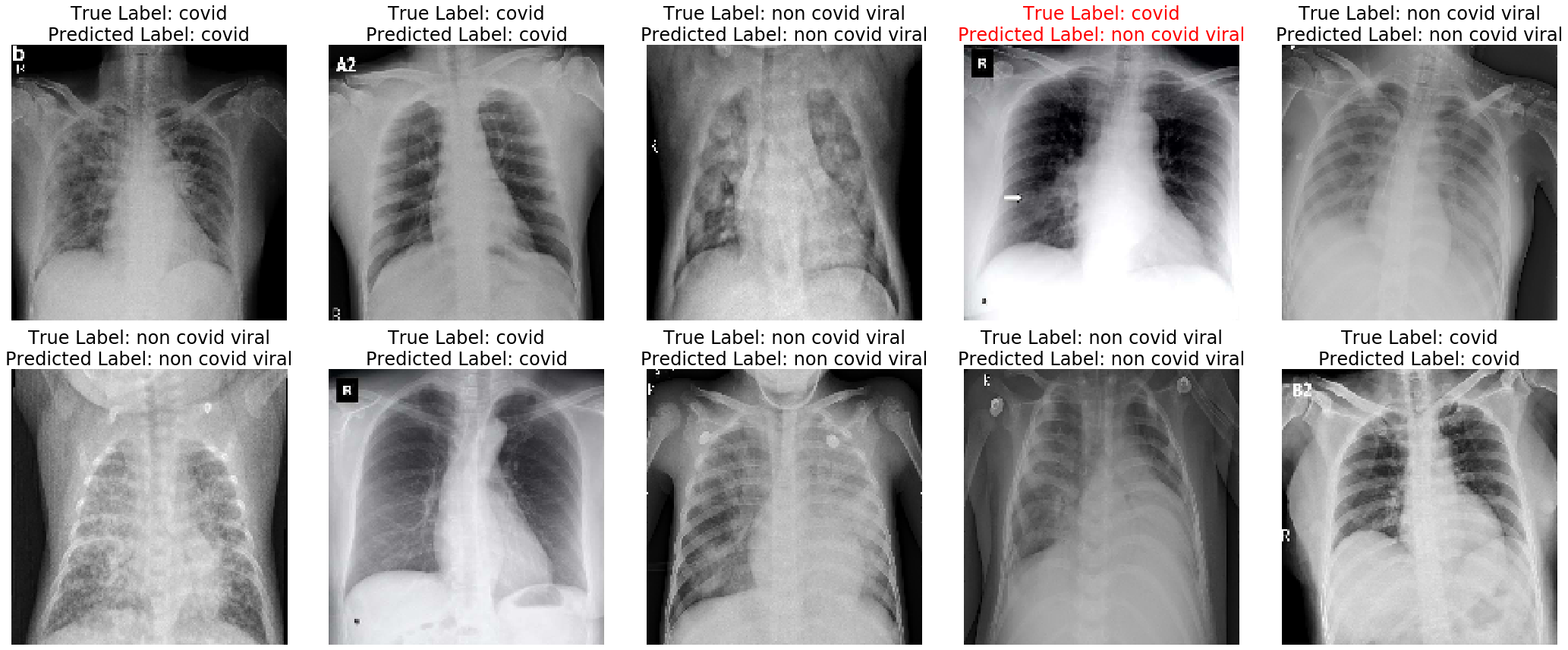
Since this third model appeared to learn the ability to distinguish COVID-19 from non COVID-19 chest X-rays, we can test this model on the test dataset. The model captured all of the COVID-19 positive samples in the test data. Below are 10 random test samples that are predicted by the model. Samples that were incorrectly predicted were labelled in red.
Conclusion
This is an interesting real-world medical imaging case study of COVID-19 detection. COVID-19 has infected nearly 2.5 million people and killed at least 171,000 worldwide, according to Johns Hopkins University. More than 42,000 people have died in the US. We looked at easy to build COVID-19 detection system leveraging AI which can give us state-of-the-art accuracy thus enabling AI for social good. As and when new COVID-19 chest X-rays are released we should be able to test improve the model. Let’s hope for more adoption of open-source AI capabilities across healthcare making it cheaper and accessible for everyone across the world!